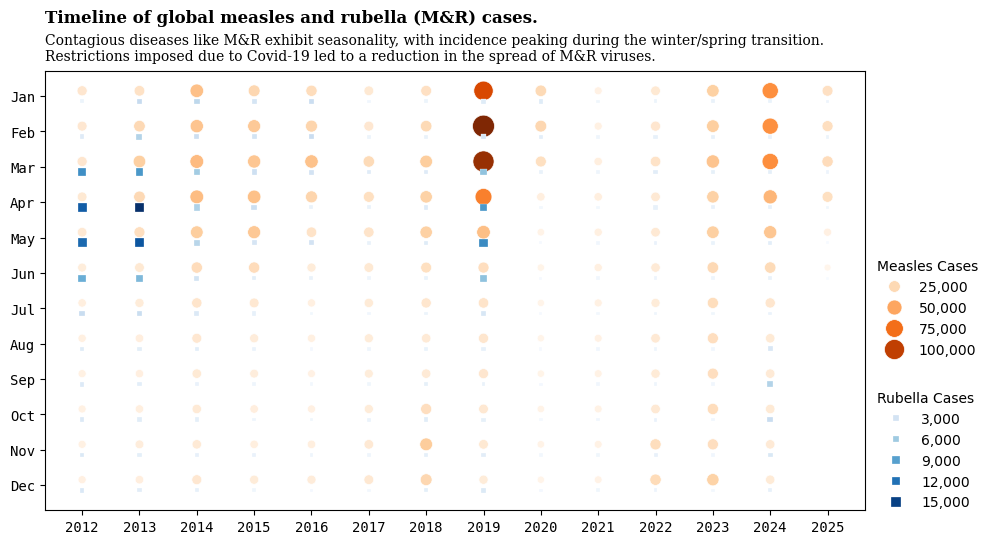
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
cases_month = pd.read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/main/data/2025/2025-06-24/cases_month.csv')
cases_year = pd.read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/main/data/2025/2025-06-24/cases_year.csv')
cases_month| region | country | iso3 | year | month | measles_suspect | measles_clinical | measles_epi_linked | measles_lab_confirmed | measles_total | rubella_clinical | rubella_epi_linked | rubella_lab_confirmed | rubella_total | discarded | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | AFR | Algeria | DZA | 2012 | 1 | 8.0 | 6.0 | 0.0 | 2.0 | 8.0 | NaN | NaN | NaN | NaN | 0.0 |
| 1 | AFR | Algeria | DZA | 2012 | 2 | 10.0 | 10.0 | 0.0 | 0.0 | 10.0 | NaN | NaN | NaN | NaN | 0.0 |
| 2 | AFR | Algeria | DZA | 2012 | 3 | 17.0 | 17.0 | 0.0 | 0.0 | 17.0 | NaN | NaN | NaN | NaN | 0.0 |
| 3 | AFR | Algeria | DZA | 2012 | 4 | 7.0 | 5.0 | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 1.0 | 1.0 | 2.0 |
| 4 | AFR | Algeria | DZA | 2012 | 5 | 14.0 | 11.0 | 0.0 | 0.0 | 11.0 | 0.0 | 0.0 | 3.0 | 3.0 | 3.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 22775 | WPR | Viet Nam | VNM | 2024 | 10 | 379.0 | 19.0 | 56.0 | 256.0 | 331.0 | 5.0 | 0.0 | 5.0 | 10.0 | 48.0 |
| 22776 | WPR | Viet Nam | VNM | 2024 | 11 | 584.0 | 37.0 | 125.0 | 347.0 | 509.0 | 0.0 | 0.0 | 1.0 | 1.0 | 75.0 |
| 22777 | WPR | Viet Nam | VNM | 2024 | 12 | 588.0 | 56.0 | 134.0 | 338.0 | 528.0 | 0.0 | 0.0 | 1.0 | 1.0 | 60.0 |
| 22778 | WPR | Viet Nam | VNM | 2025 | 1 | 156.0 | 7.0 | 0.0 | 124.0 | 131.0 | NaN | NaN | NaN | NaN | 25.0 |
| 22779 | WPR | Viet Nam | VNM | 2025 | 2 | 22.0 | 0.0 | 0.0 | 20.0 | 20.0 | NaN | NaN | NaN | NaN | 2.0 |
22780 rows × 15 columns